A fct that plot a beautiful matrix from an sbm object or a network matrix it does have suitable parameters to get the wanted plots. This is the generic function: it does have one method Bipartite and one for Simple Sbm. The `x` object need to be construct by one of the `estimate***SBM` function from the `sbm` package.
Usage
plotSbm(
x,
ordered = FALSE,
transpose = FALSE,
labels = NULL,
plotOptions = list()
)Arguments
- x
Sbm model of class `BipartiteSBM_fit`, `SimpleSBM_fit` or simple numeric `matrix`.
- ordered
Boolean. Set
TRUEif the matrix should be reordered (Default isFALSE)- transpose
Boolean. Set
TRUEto invert columns and rows to flatten a long matrix (Default isFALSE)- labels
a named list (names should be: `"col"` and `"row"`) of characters describing columns and rows component (Default is
NULL)- plotOptions
a list providing options. See details below.
Value
a ggplot object corresponding to the matrix plot inside the app. Groups the network matrix is organized by blocks, the small tiles are for individuals connections. The big tiles between red lines are for block connectivity
Details
The list of parameters plotOptions for the matrix plot is
"showValues": Boolean. Set TRUE to see the real values. Default value is TRUE
"showPredictions": Boolean. Set TRUE to see the predicted values. Default value is TRUE
"title": Title in characters. Will be printed at the bottom of the matrix. Default value is NULL
"colPred": Color of the predicted values, the small values will be more transparent. Default value is "red"
"colValue": Color of the real values, the small values will close to white. Default value is "black"
"showLegend": Should a legend be printed ? TRUE or FALSE, default: FALSE
"interactionName": Name of connection in legend default: "Connection"
Examples
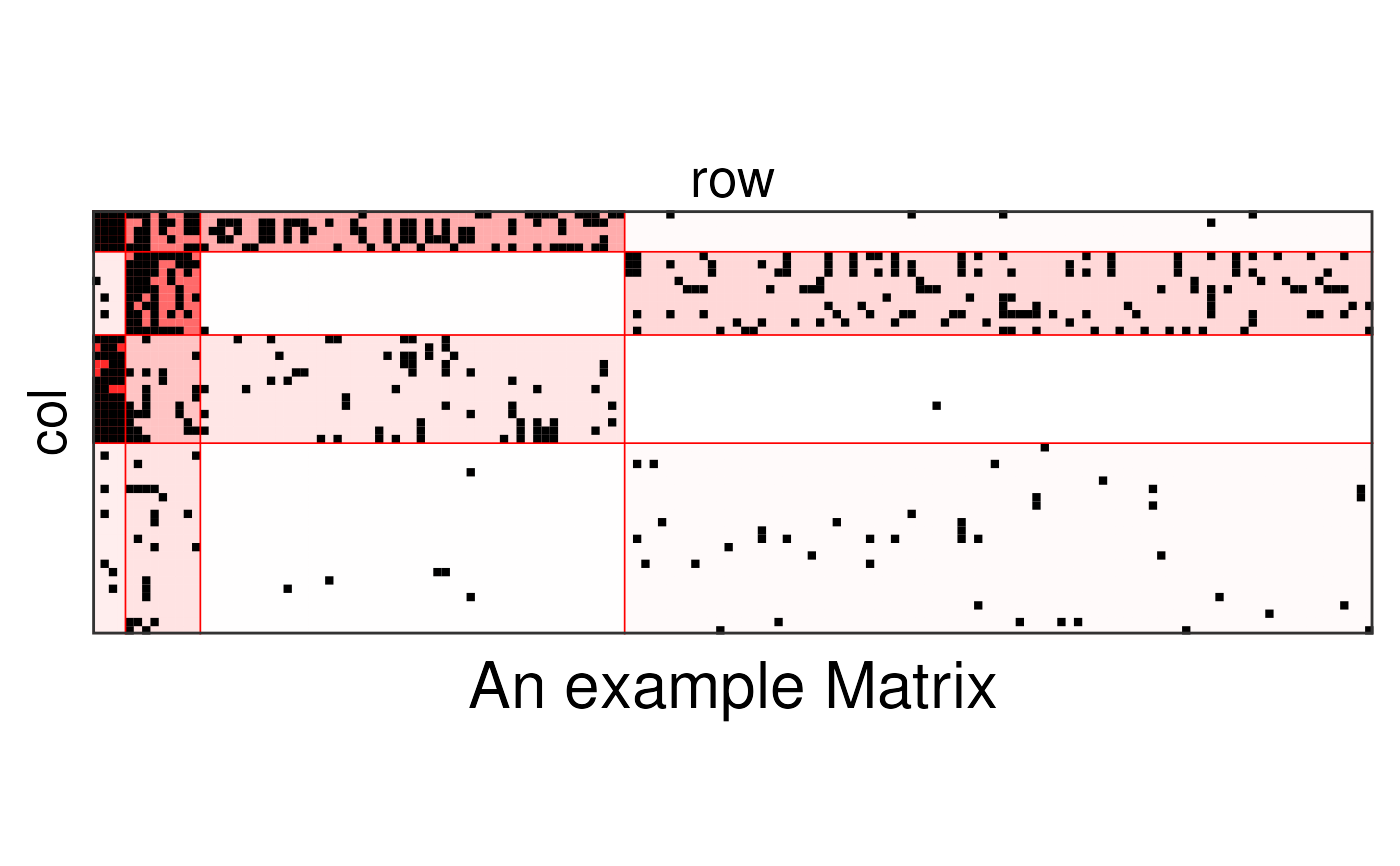
# my_sbm_bi <- sbm::estimateBipartiteSBM(sbm::fungusTreeNetwork$fungus_tree,
# model = 'bernoulli')
my_sbm_bi <- FungusTreeNetwork$sbmResults$fungus_tree
plotSbm(my_sbm_bi,
ordered = TRUE, transpose = TRUE,
plotOptions = list(title = "An example Matrix")
)
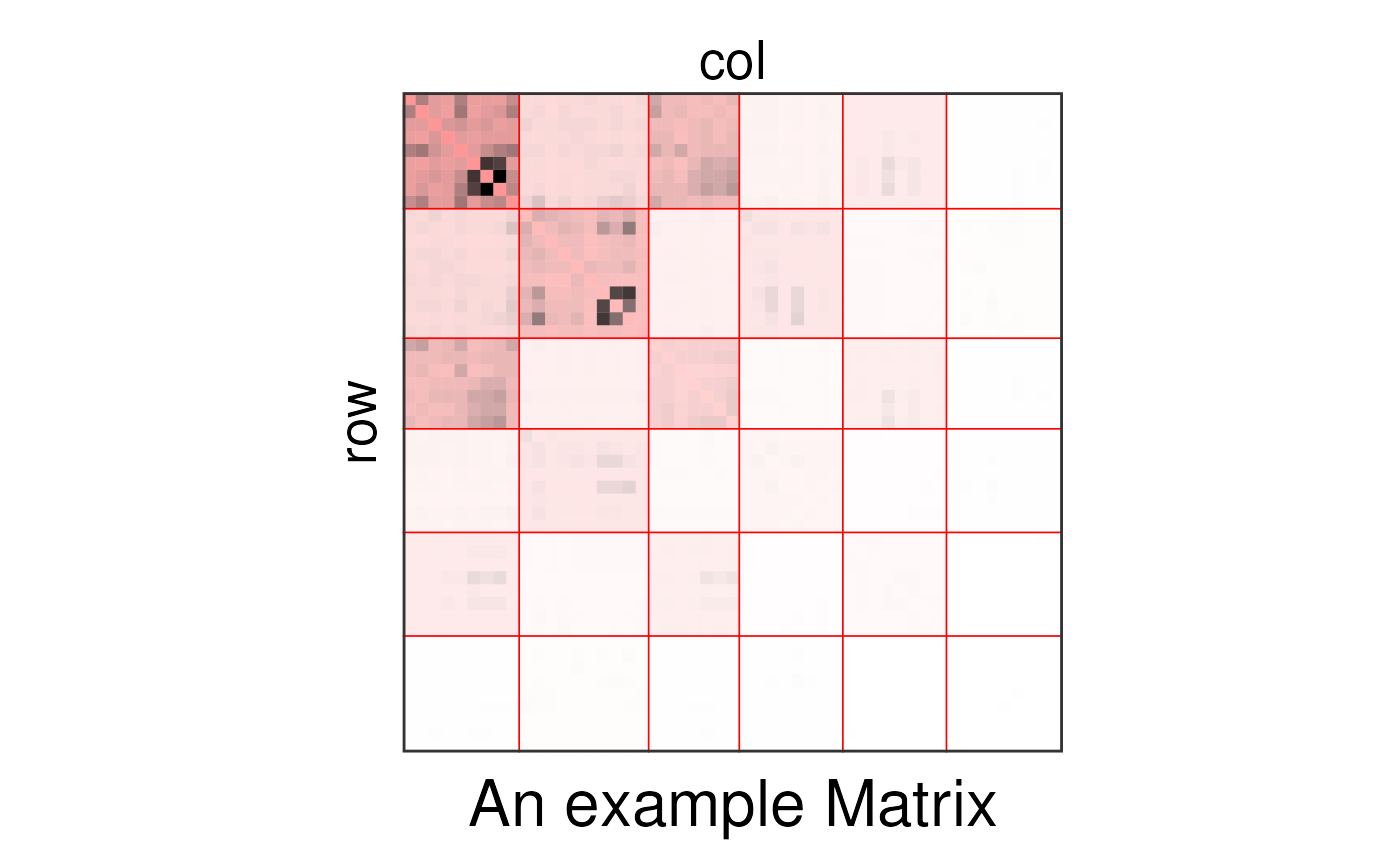
 # my_sbm_uni <- sbm::estimateSimpleSBM(sbm::fungusTreeNetwork$tree_tree,
# model = "poisson")
my_sbm_uni <- FungusTreeNetwork$sbmResults$tree_tree
plotSbm(my_sbm_uni,
ordered = TRUE,
plotOptions = list(title = "An example Matrix")
)
# my_sbm_uni <- sbm::estimateSimpleSBM(sbm::fungusTreeNetwork$tree_tree,
# model = "poisson")
my_sbm_uni <- FungusTreeNetwork$sbmResults$tree_tree
plotSbm(my_sbm_uni,
ordered = TRUE,
plotOptions = list(title = "An example Matrix")
)
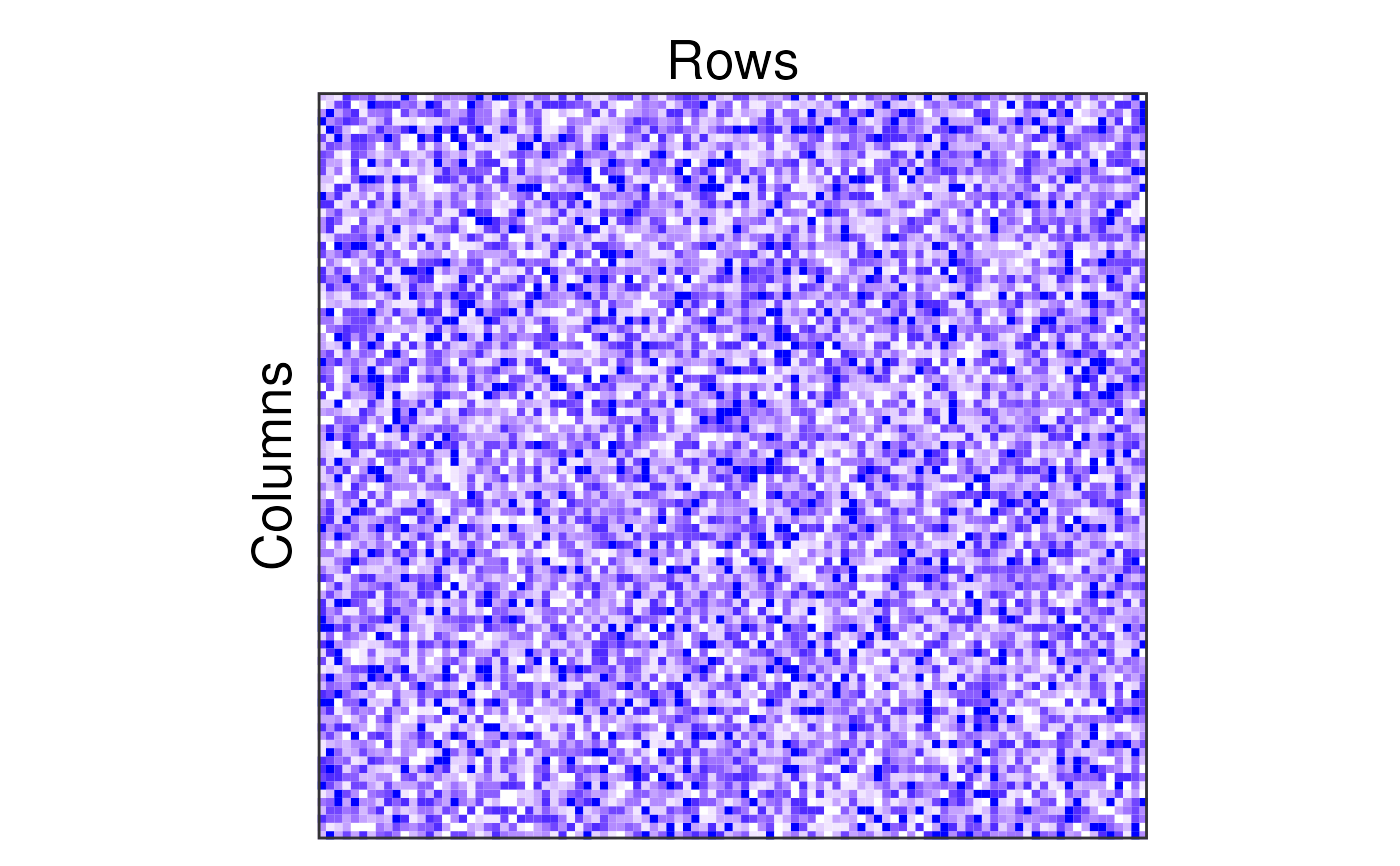
 n_col <- 100
n_row <- 90
mat <- matrix(sample(0:10, n_col * n_row, replace = TRUE), n_col, n_row)
plotSbm(mat,
transpose = TRUE,
labels = list(col = "Columns", row = "Rows"),
plotOptions = list(colValue = "blue")
)
n_col <- 100
n_row <- 90
mat <- matrix(sample(0:10, n_col * n_row, replace = TRUE), n_col, n_row)
plotSbm(mat,
transpose = TRUE,
labels = list(col = "Columns", row = "Rows"),
plotOptions = list(colValue = "blue")
)